Visualization of the clonal distribution as a function of the rank
Source:R/plotFunctions.R
plotRankDistrib.RdThis function plots the clonal distribution, at any repertoire level, as a function of the occurrence rank within each sample.
For instance, the most frequent sequence is attributed a rank of 1, and its relative abundance is plotted on the y-axis.
Arguments
- x
an object of class
RepSeqExperiment- level
a character specifying the level of the repertoire to be taken into account to plot the clonal distribution. Should be one of "aaClone", "ntClone", "ntCDR3" or "aaCDR3".
- scale
a character specifying whether to plot the clonal abundance in "count" or "frequency" or "log" scale.
- ranks
an integer specifying the number of top ranks to be plotted. Default is NULL, which plots all ranks.
- grouped
a boolean indicating whether or not the mean and se of samples belonging to the same experimental group specified in the ColorBy parameter should be computed. Grouping is performed on the chosen groupe in colorBy. Default is FALSE.
- colorBy
a character indicating one column name in mData. Colors are thus attributed to the different groups within this column. The chosen column must be of class factor.
- facetBy
a vector of character indicating one or two column names in mData to apply a facet on.
- label_colors
a list of colors for each variable in ColorBy. See
plotColors. If NULL, default colors are used.
Examples
data(RepSeqData)
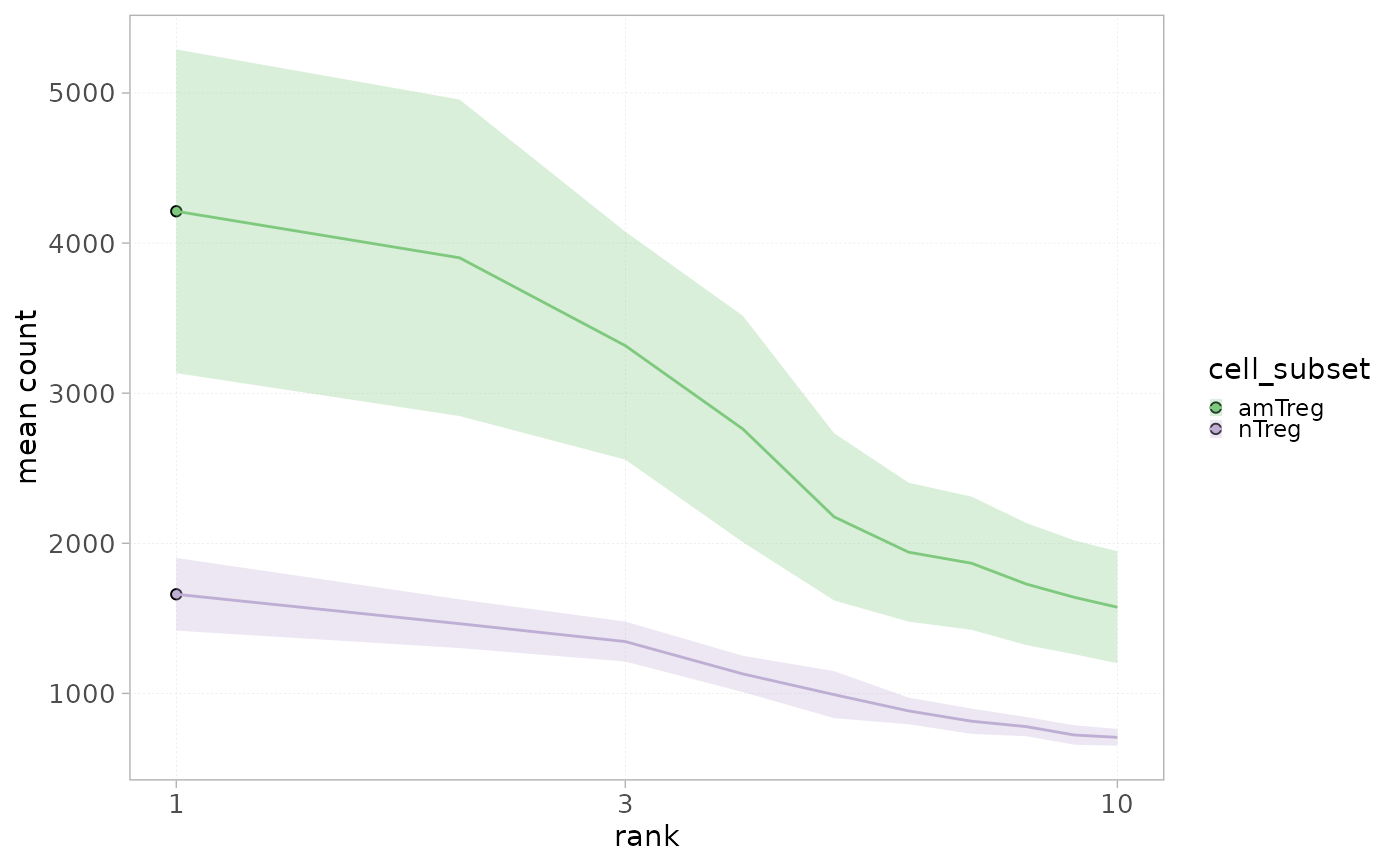
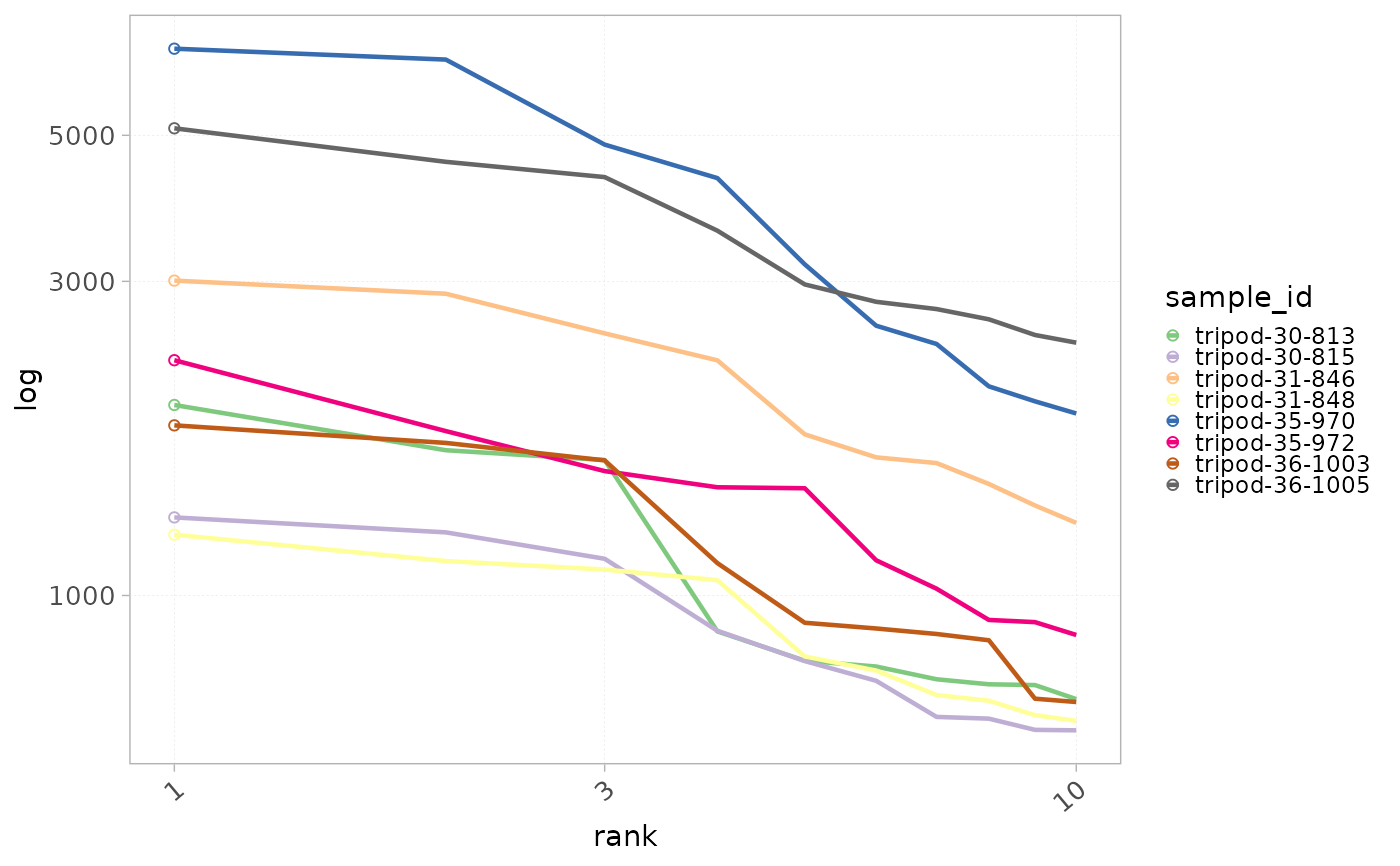
plotRankDistrib(x=RepSeqData, level = "aaCDR3",
scale="log",
ranks=10,
colorBy="sample_id" )
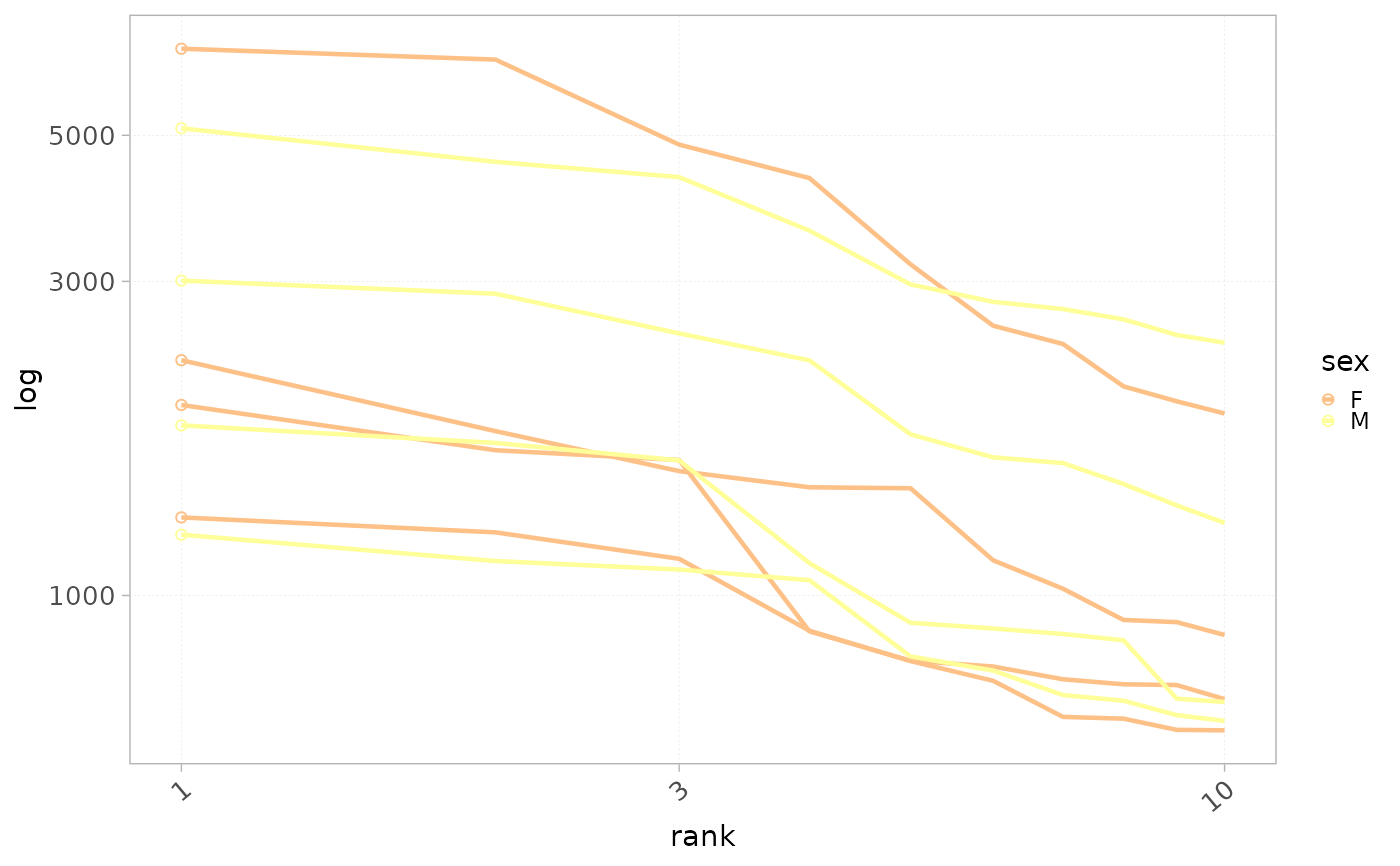
 plotRankDistrib(x=RepSeqData, level = "aaCDR3",
scale="log",
ranks=10,
colorBy="sex" )
plotRankDistrib(x=RepSeqData, level = "aaCDR3",
scale="log",
ranks=10,
colorBy="sex" )
 plotRankDistrib(x=RepSeqData, level = "aaCDR3",
scale="count",
ranks=10,
colorBy="cell_subset",
grouped=TRUE )
plotRankDistrib(x=RepSeqData, level = "aaCDR3",
scale="count",
ranks=10,
colorBy="cell_subset",
grouped=TRUE )