Down-sampling
This strategy can be applied when the studied samples largely differ in their repertoire sizes.
By using the sampleRepSeqExp() function, users can
choose the value to which all the samples are downsampled. If not
specified, the lowest number of sequences across the dataset will be
used.
This function returns a new RepSeqExperiment object with the downsized data.
RepSeqData_ds<- sampleRepSeqExp(x = RepSeqData,
sample.size = 50000)
#> You set `rngseed` to FALSE. Make sure you've set & saved
#> the random seed of your session for reproducibility.
#> See `?set.seed`
#> Down-sampling to 50000 sequences...
#> Creating a RepSeqExperiment object...
#> Done.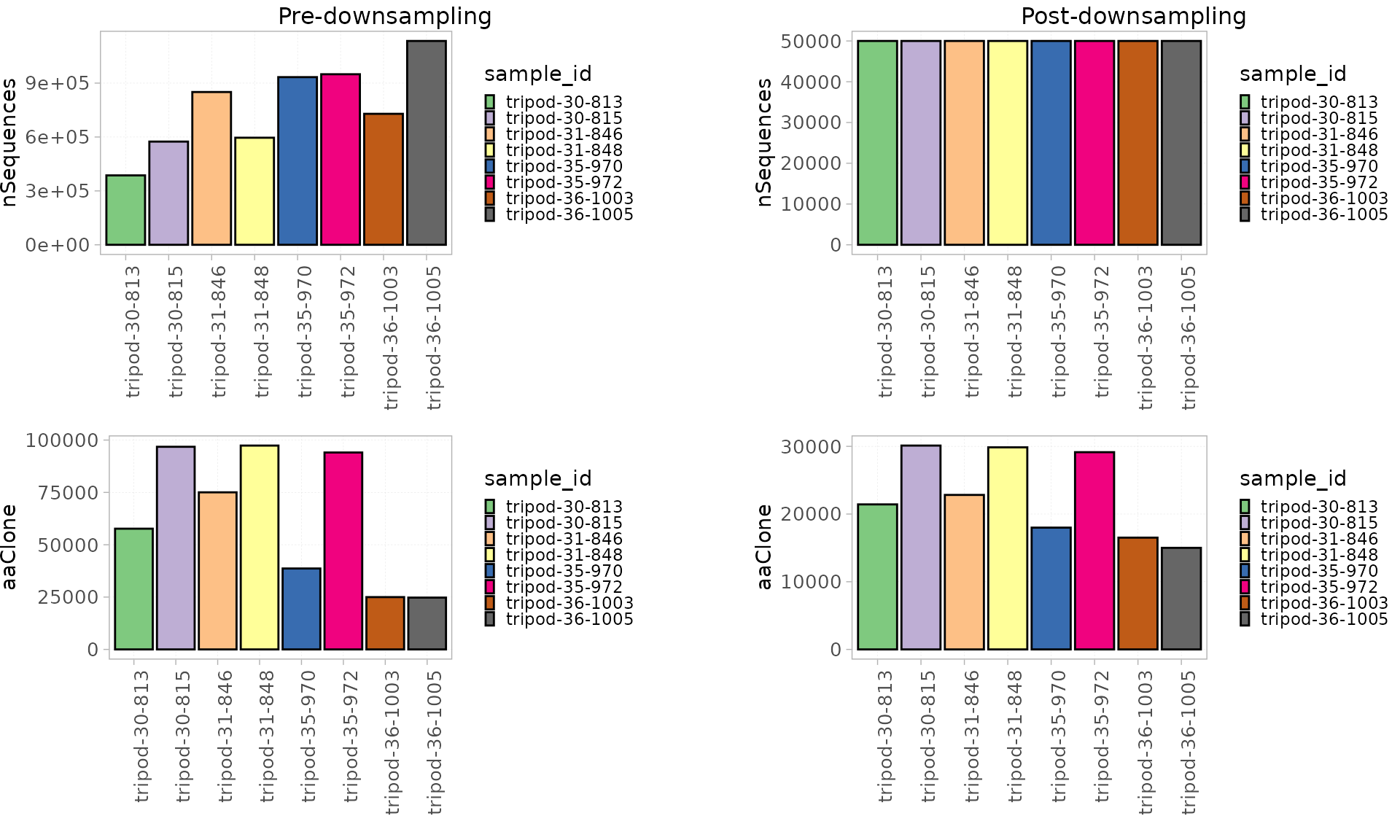
Shannon-based normalization
This strategy adapted from Chaara et al., 2018
can be used to eliminate “uninformative” sequences resulting from
experimental noise. It uses the Shannon entropy as a threshold and is
applied at the ntClone level. This strategy is particularly
efficient when applied on small samples as it corrects altered count
distributions caused by a high-sequencing depth.
The function ShannonNorm() allows the application of
this strategy without the need to specify any parameter and returns a
new RepSeqExperiment object with the corrected data.
RepSeqData_sh <- ShannonNorm(x = RepSeqData)
#> Creating a RepSeqExperiment object...
#> Done.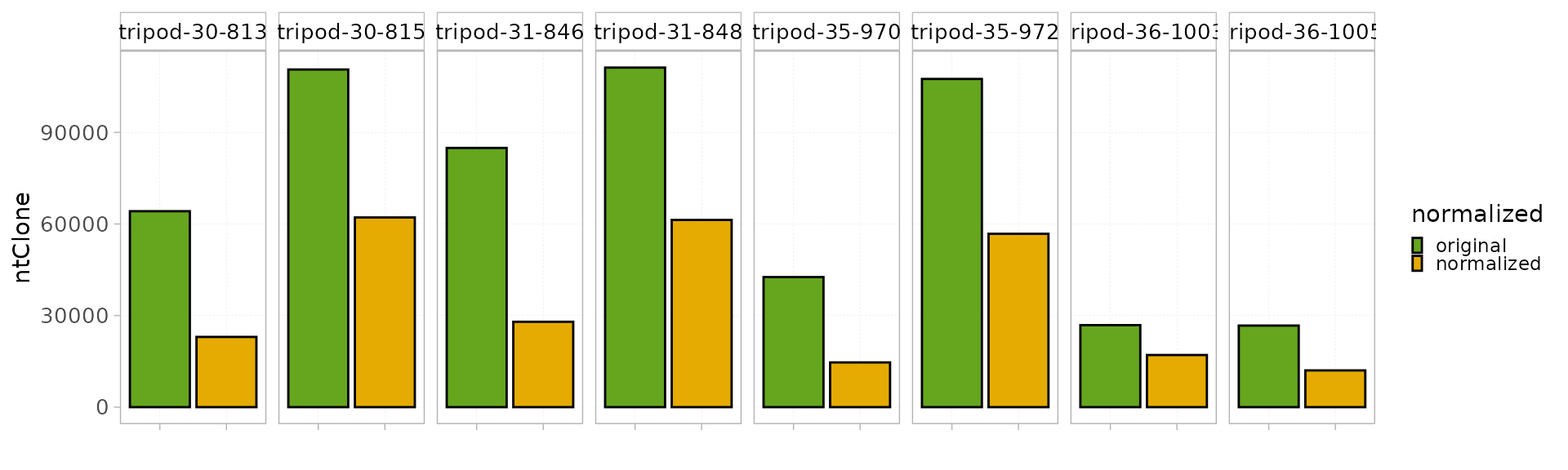
Notes:
- Eliminated sequences in each sample are stored in the otherData slot.
- Chao, the richness estimator, isn’t recalculated for normalized datasets as as their original composition has been modified.
